
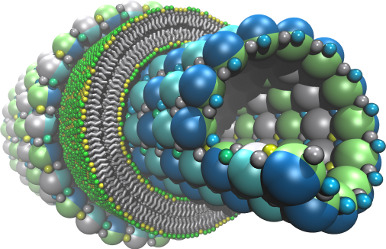
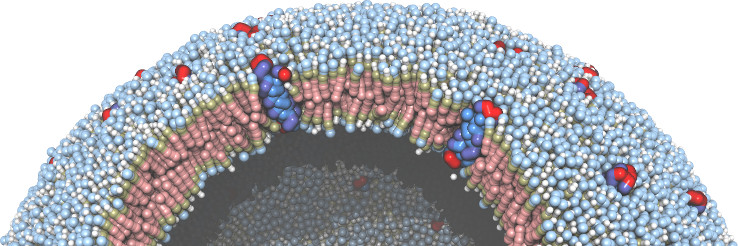





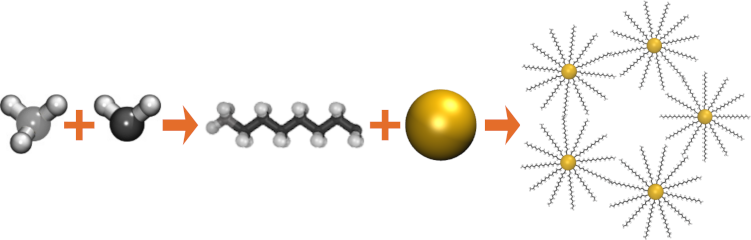


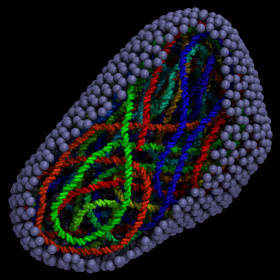













 (built with moltemplate;
visualized with
VMD,
OVITO,
and
topotools
)
(built with moltemplate;
visualized with
VMD,
OVITO,
and
topotools
)
DESCRIPTION
LAMMPS is an extremely flexible and customizable molecular dynamics engine. Moltemplate is a general cross-platform text-based molecule builder for LAMMPS. Moltemplate was intended for building custom coarse-grained molecular models, but it can be used to prepare realistic all-atom simulations as well. It supports the ATB molecule database, as well as a wide variety of existing force fields and models including: OPLSAA (2023) , OPLSUA (2008) , LOPLS (2015) , AMBER (GAFF,GAFF2) , DREIDING , COMPASS , MOLC , TraPPE (1998) , EFF , mW , ELBA (water) , and oxDNA2 . But it can also be used to build molecules using any of the exotic force fields (and atom styles) available in LAMMPS, including new force fields created by modifying the LAMMPS source code. (Note: Careful selection of atom types is necessary. Moltemplate does not support atom typing, and is not suitable for all-atom protein simulations.) Molecules can be copied, combined, and linked together as building-blocks to define new molecules (hierarchically). Once built, individual molecules and subunits can be customized (atoms, bonds, and subunits can be moved and deleted). Moltemplate is inter-operable with ATB , LigParGen , AmberTools , OpenBabel , RED-server , VMD , topotools , PACKMOL , EMC , CellPACK , Vipster , struc2lammpsdf , and any other program that generates MOL2 or LAMMPS DATA (.lmpdat) files (by using the mol22lt.py and ltemplify.py file converters).
COARSE-GRAINED MODELING
Coarse-grained molecular models often require more complex and unconventional force fields, compared to traditional all-atom models. The LAMMPS and MOLTEMPLATE programs allow users extraordinary freedom to customize the forces acting on the atoms in a molecule. Users can also combine radically different molecular representations together including point-like, rigid, and continuum-field/atomistic hybrids. The LAMMPS code is open-source and written in a modular style so that new force fields and features can be easily added by end users. (And moltemplate is written in a style to easily accommodate these changes.)| Unlimited levels of composition make it easy to build complex hierarchical assemblies frequently encountered in biology. |

|
USAGE
moltemplate.sh [-atomstyle style] [-pdb/-xyz coord_file] [-a assignments.txt] [-vmd] file.lt
TYPICAL USAGE:
An LT-file containing molecule definitions can be converted to LAMMPS input files using moltemplate.sh:
moltemplate.sh system.ltor
moltemplate.sh -xyz coords.xyz -atomstyle "full" -vmd system.lt
Either of these commands will construct a LAMMPS data file and a LAMMPS input script (and possibly one or more auxiliary input files), which can be directly run in LAMMPS with minimal editing.
In the first example, the coordinates of the atoms in the system are built from commands inside the "system.lt" file. In the second example coordinates for the atoms are read from an XYZ-file, and then VMD is invoked to visualize the system just created. (PDB-files and simple 3-column coordinate files are also supported. The "full" atom style was used in this example, which is also the default. However other LAMMPS atom styles are supported, including hybrid style-lists, enclosed in quotes.)
Existing LAMMPS input/data files can be converted into moltemplate (".LT") format using the "ltemplify.py" utility.
Moltemplate requires the Bourne-shell, and a recent version of python
(2.7 or 3.0 or higher), and can run on OS X, linux, or windows
(if a suitable
shell environment has been installed).
Substantial memory required.
(Between 3 and 12 GB of RAM needed per 1000000 atoms.)
LICENSE
Moltemplate is free and is available under the terms of the open-source
MIT
and
PSF
licenses.
(Details here. )
CITATION
If you find this program useful, please cite:
"Moltemplate: A Tool for Coarse-Grained Modeling of Complex Biological Matter and Soft Condensed Matter Physics", J. Mol. Biol., 2021, 433(11):166841, Jewett AI, Stelter D, Lambert J, Saladi SM, Roscioni OM; Ricci M, Autin L, Maritan M, Bashusqeh SM, Keyes T, Dame RT; Shea J-E, Jensen GJ, Goodsell DS
https://doi.org/10.1016/j.jmb.2021.166841
November 16th, 2025
Andrew Jewett
