About the title image ...
This structural model shown in the image below is a model of a
lipid protein nanotube ,
(LPN):
U. Raviv, D.J. Needleman, Y. Li, H.P. Miller, L. Wilson, and C.R. Safinya,
PNAS, 102(32), 11167-11172, 2005
This is an example how to define molecule objects hierarchically
in moltemplate...
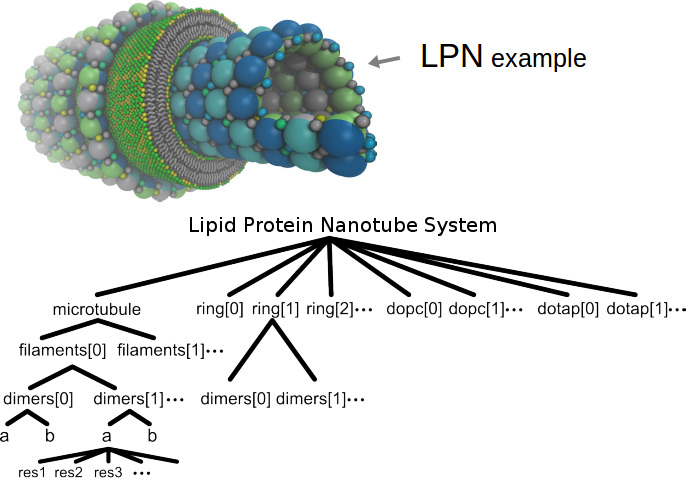
In this example,
tubulin
monomers (named "a" and "b", shown partly in dark-blue and light-blue) were built from smaller subunits (in this case, a handful of irregularly sized particles with various colors), bonded together to define "dimers". Dimer objects can be used as building blocks to define even larger objects (named "filaments" and "rings"), which in turn can be used to define
microtubules.
Arrays of coarse-grained "dopc" and "dotap" lipids were then arranged around the microtubule using simple "move()" and "rot()" commands to form a tube-shaped bilayer, as explained in
this talk (slides 106-127).
(Defining objects hierarchically is optional. If you prefer, you can simply create a 2-D array of dimers, or a single long list of monomers.
The force-field parameters for this example was never tuned to reproduce
realistic self-assembly behavior.
3-D graphics generated by
VMD
&
topotools . (High
resolution
version.)
November 21 2016
Andrew Jewett 


